Project Introduction and Planning
Project Introduction and Planning
This section is happening at the end of Thursday
Now you have reached the final module of the course, where you will apply what you learn in this course on a new problem.
We have provided a dataset for you to perform the analysis as a group. We recommend that you work individually on your computer (so that you get your own practice), and share with the group when you hit an issue, or when you find something interesting.
In the end, the group will work together to produce one group presentation. Each group will then have 8-10 minutes to present their analysis and their answers to the project questions.
It could be helpful to have a group workspace on a shared document (such as Google Doc, Google Jamboard) where your group can share resources and ideas during the course, or even draw a workflow. Google Slide can be useful for co-creating group slides for the presentation.
Even though it’s the same dataset, it is always interesting to see how people may come up with different ways of looking at the data, variations of genes of interest, and go down different path for downstream analysis.
The dataset for group project is…
Brugia malayi male and female of different stages
Here we have RNA-seq of B. malayi from larval stages to various ages of male and female from Grote et al. (2017). In their paper, Grote et al. investigated the relationship between B.malayi and their symbiont bacteria Wolbachia, but they did not explain much about the gene expression changes of the worms in the different life cycle stages of the parasite. There are various pair-wise comparisons we could do here. Also, if you are intested in dual-RNA-seq, we also provide the FASTQ files that will have sequencing reads from the Wolbachia. You could use these FASTQ files to do the mapping to a relevant Wolbachia genome.
Key questions
- What happened to male and female worms as they develop into adults?
- At which point do gene expression in male and female worms become very different?
- Challenge question: If you want to do dual-RNA-seq analysis, starting from the raw FASTQ file from a sequencing platform, what are the steps in the workflow that need to be done differently (e.g. different tools, different genome resources, etc)?
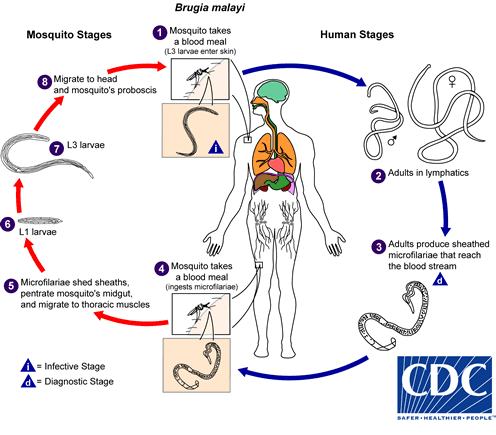
Figure 4.1: Brugia malayi life cycle
PS. The questions, especially the Challenge questions, may not have exactly “the correct answer” but they are to allow discussion within groups, and with instructors. Plus, perhaps chances to reflex and discuss their current/future genomics projects