Introduction
Introduction to module 3C
Genomic surveillance of pathogens aims to understand the emergence and dissemination of pathogens or their lineages of risk with the ultimate goal of implementing evidence-based interventions to protect public health. Effective, real-time genomic surveillance relies on the streamlined collection, analysis and integration of diverse sources of data.
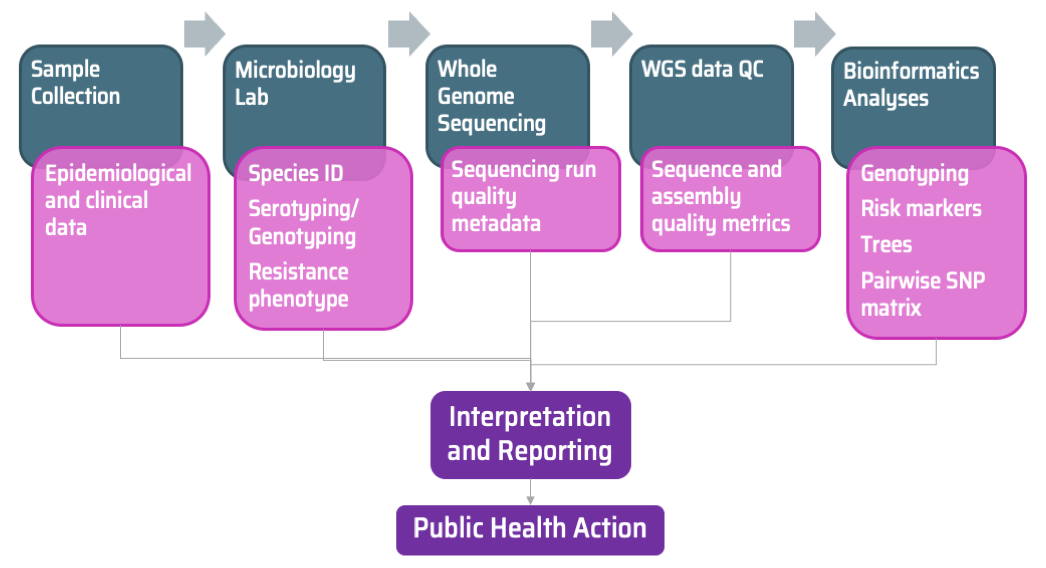
Module 3A reviewed the fundamentals of specimen collection, a process during which epidemiological data from patients is also collected by healthcare professionals. Data on species identification and phenotypic or molecular characterization of the isolates is often generated by the reference laboratory or by microbiology laboratories linked to healthcare facilities. In an ideal scenario, the different sources of laboratory data are stored in a centralised surveillance system and database (such as WHONET).
Module 3B reviewed how the quality of the material to be sequenced, and that of the sequence data itself may alter how data can be interpreted. Bioinformaticians often use different analytic tools to extract useful data from the genomes, such as presence of markers of risk (e.g., AMR and virulence genes), genotyping information (e.g., MLST, cgMLST, and other species-specific typing), and phylogenetic relationships (trees), as well as quality metrics. Module 1B summarised some of the analytic tools and resources commonly used for data analysis. However, these outputs are rarely incorporated into the same database as epidemiological and laboratory data.
The genomic epidemiologist’s job often starts by combining data from diverse sources that might not be complete or standardised to facilitate the identification of relevant patterns for meaningful interpretation and intervention. The data and any patterns often need to be digested and summarised for decision makers. To this purpose, the Centre for Genomic Pathogen Surveillance develops free web applications for integration and visualisation of surveillance (and other) data, called data-flo and Microreact.
The aim of this module is to highlight the role of different analytics and sources of data in pathogen surveillance, and the value of streamlined data integration for real-time decision making, while also introducing resources that can facilitate the inclusion of this topic in teaching curricula.
Learning Outcomes
At the end of this module participants will be able to:
Identify knowledge, concepts and skills required for data analysis and integration, such as data sources, formats, standards, and data merging.
Determine appropriate content for designing training in data analysis and integration.
Apply pedagogical principles to learning strategies, such as adult learning and active learning.
Design relevant activities for training in data analysis and integration, such as use cases and group work.
Provide an outline of tools and resources needed to deliver training in data analysis and integration.
Assess knowledge and skills related to data analysis and integration, such as through group discussion.
Presentation delivered by the instructor (Silvia Argimón) during the session.